The experiment
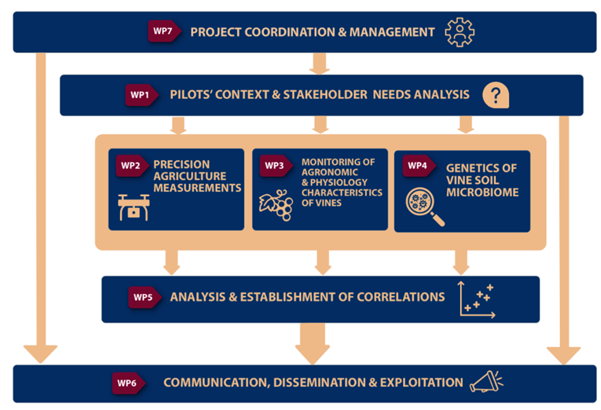
To set the DiVine methodology into action, a total project duration of 36 months with 7 Work Packages is foreseen and structured to allow active planning and management for achieving the project’s goals. There are four technical Work Packages (WP2 to WP5) involving R&D and innovation activities with continuous progress assessment between the individual scientific and methodological approaches. WP6 will focus upon Communication, Dissemination and Exploitation activities. In addition to that, the project will be carried out under the governance of WP7, which will be dedicated to the management and evaluation of the project’s progress and results (Figure 2). The ten milestones proposed will serve as a benchmark ensuring the delivery of all the WP-related results. Risk assessment is also in place to proactively apply corrective measures, in the case that problems arise, and facilitate a smooth project implementation.
Research in DiVine will be conducted as part of a design revolving around the combined application of PA measurements, agronomical and physiological examination of the plant and soil tissues, and molecular analysis of the soil microbiome. All research activities will take place in the AUA experimental vineyard field at Spata, Attica (22 ha with Greek varieties) and will be scaled-up in two commercial vineyards in the same region. The main steps of the proposed multi‐level methodology involve:
- Imprinting the agronomic and physiology characteristics
ELGO-Demeter, the organisation collaborating in the project, will monitor the agronomic and physiology characteristics of the vines, the soil properties and nutrient condition, and the plant nutrient condition by conducting plant tissue analysis. Moreover, a number of weed-related indices are going to be measured and associated with measurements of the crop phenotype and the soil microbiome. Finally, a number of organoleptic properties will be also measured together with the estimation of yield and quality at harvest using grid yield and quality estimation. - Densely sampling soil and analysing the genetic and epigenetic profile of the samples with simultaneous application
ELGO-Demeter in collaboration with the Genetics Lab team of AUA will carry out the sampling of the soil microbiome with the use of in-house developed lysimeters, which have the ability to reliably collect microbiome from the unsaturated zone of the cultivation, and to depths of interest, in a cost-efficient way. The genetic and molecular analysis of the samples will include next generation sequencing (NGS) methods that will identify the species of the soil microorganisms by examining the bases sequence of the 16S rRNA gene, as this region is unique to each microorganism. This will contribute to the genetic and evolutionary identification of the soil microbiota. Proteomic and metabolic assays will follow for the qualitative and quantitative evaluation of protein content in a given environment and the epigenetic profiling of the collected samples. These technologies have emerged as a powerful tool for investigating the structural, evolutionary and metabolic properties of complex microbial communities. The microbial activity of the grapevine will be also monitored by macroscopic surveillance, as the metabolic processes that occur in microorganisms result in heat production. The Infrared Thermography (IRT), using UAVs with multispectral cameras, will allow the detection and monitoring of patterns of microbial activity on a large scale of samples without affecting the soil. In parallel, the use of remote sensing tools for continuous monitoring of ground and air conditions will complement the acquired dataset for microbiome imprinting. The quality of a region’s microbiota changes over time depending on the climate as the soil profile alters. Therefore, screening of environmental conditions will allow the monitoring, verification and prediction of qualitative and quantitative changes in the soil microbiome. - Geostatistical analysis, correlations, and cross validation
The core challenge of DiVine is to establish correlations between the soil microbiome distribution and activity, and the resulting phenotypic traits of grapevines. To this end, at the final stage of the project, the three datasets collected during each of the three growing seasons, will be combinedly analysed with the aim to investigate correlations and explore how the parameters collected and monitored both at the macro- and microscopic level may affect the quality and quantity of vines. The datasets available, including PA measurements, soil microbiome data, and agronomic and physiology measurements will be the input to geostatistical analysis and cross-validation, modelling, and deep learning algorithms deployment. The geostatistical analysis will provide the basis for the development of the final yield model, while deep learning methods are going to be employed for cross-validation purposes due to the high volume of the data to be retrieved throughout the project.